Estimate the model#
Load the data#
We can use the load_descriptors() and
load_retention_times() convenience functions that can load
data from tab separated values files provided that:
the retention times are in the first column, and
the molecular descriptors are in the last columns and follow at least one non numeric column (such as, for instance, the SMILES one).
from jp2rt import load_descriptors, load_retention_times
X = load_descriptors('plasma+descriptors.tsv')
y = load_retention_times('plasma+descriptors.tsv')
As a check we can verify that the number of rows in X is the same as the
number of rows in y:
X.shape, y.shape
((799, 243), (799,))
Use scikit-learn#
To understand more in detail how model estimation and prediction works, in this
section we will use directly the scikit-learn
package using the regression models of the sklearn.ensemble package; in
the following section we’ll present an easier approach based on jp²rt that
summons the approach shown here.
Clean the data#
Before using the data we need to prepare it as detailed in Preprocessing data.
The computation of the descriptors may have produced some NaNs, we’ll follow two approaches according to the fact that we have columns with just NaNs, or with some NaNs and other valid values. First of all, we compute the columns containing just NaNs:
import numpy as np
all_nan_cols = [i for i, x in enumerate(X.T) if all(np.isnan(x))]
all_nan_cols
[216]
We can take care of such columns using a
ColumnTransformer that will drop the columns
containing just NaNs (and pass through the other ones); the case of some NaNs
will be handled using a SimpleImputer that will replace
missing values with the mean of valid ones; moreover we’ll scale the data using
a StandardScaler to reduce the variance, a fact
that is usually beneficial to regression models.
from sklearn.compose import ColumnTransformer
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import StandardScaler
from sklearn.pipeline import make_pipeline
drop_all_nan_cols = ColumnTransformer([('drop_all_nan_cols', 'drop', all_nan_cols)], remainder = 'passthrough')
simple_imputer = SimpleImputer()
standard_scaler = StandardScaler()
We’ll put the column transformer, the imputer, and the scaler together with
various regressors (discussed in the coming subsections) in a pipeline using
the make_pipeline() function.
Model selection#
To decide with ensemble model will best fit our data we’ll use both cross
validation and
visual
inspection
of the results via the convenience jp²rt function
evaluate_model() that returns a dictionary with various
information of interest.
Gradient Boosting#
Let’s start defining the model pipeline based on the
GradientBoostingRegressor and evaluating it:
from sklearn.ensemble import GradientBoostingRegressor
from jp2rt import evaluate_model
regressor = GradientBoostingRegressor()
model = make_pipeline(drop_all_nan_cols, simple_imputer, standard_scaler, regressor)
evaluation = evaluate_model(model, X, y)
We keep track of this first evaluation in the results list, and we can inspect
the plot key of evaluation
result = []
result.append({'regressor': 'GradientBoosting'} | evaluation['metrics'])
evaluation['plot']
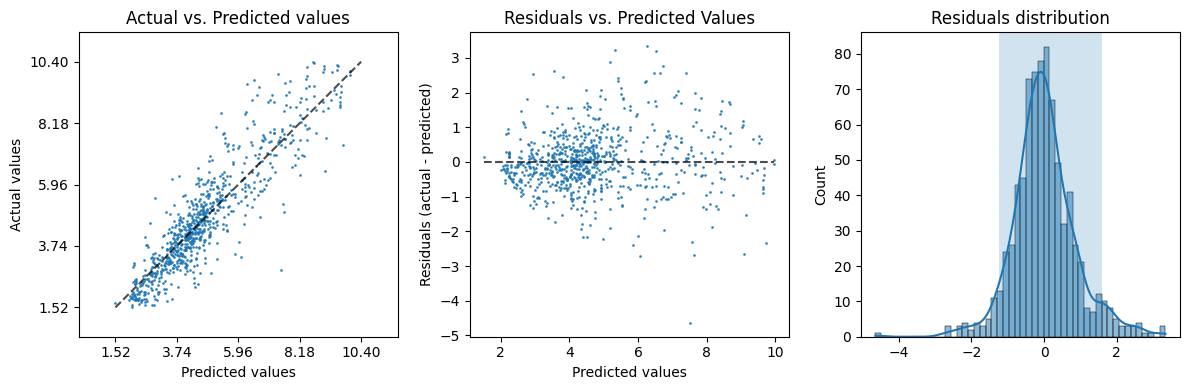
that depicts a some residuals (the difference between the true and the
predicted values) analysis; in a more quantitative way we can inspect the
metrics key, or the convenience version table:
print(evaluation['table'])
+-----------+---------+
| r2 mean | 0.8028 |
| r2 std | 0.0333 |
| rmse mean | 0.8617 |
| rmse std | 0.0931 |
| q0 | -1.2509 |
| q1 | 1.5986 |
+-----------+---------+
Since the cross validation runs a model estimation per split (in this example, 5 times, that is the default value for the evaluation function), we get the mean and std (standard deviation) of two metrics of interest:
the coefficient of determination (\(R^2\)) computed by
r2_score(), andthe root mean squared error (RMSE) computed by
root_mean_squared_error().
Moreover, the evaluation returns the values of the empirical quantiles of the
residual distribution (obtained using the mquantiles()
function) \(q_0\) at 5% and \(q_1\) at 95%. The interval \([q_0, q_1]\) is the shaded
box in the last of the three plots and represents the 90% of the residuals
according to the cross validated prediction (obtained using the
cross_val_predict() function).
A more robust boosting model#
The HistGradientBoostingRegressor is a more recent
implementation of the gradient boosting algorithm that is more efficient and can
handle larger datasets, taking care of NaNs; we can use it with a simpler
pipeline:
from sklearn.ensemble import HistGradientBoostingRegressor
regressor = HistGradientBoostingRegressor()
model = make_pipeline(drop_all_nan_cols, regressor)
evaluation = evaluate_model(model, X, y)
result.append({'regressor': 'HistGradientBoosting'} | evaluation['metrics'])
evaluation['plot']
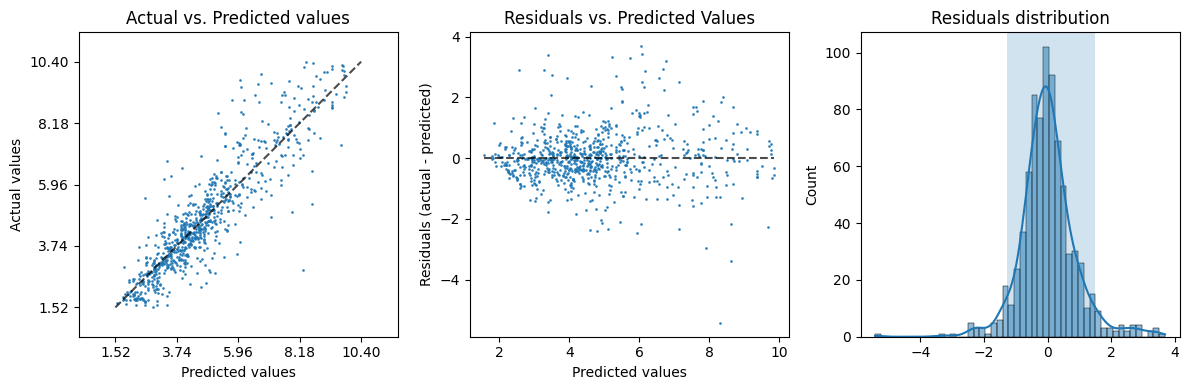
Quantitatively the results look promising:
print(evaluation['table'])
+-----------+---------+
| r2 mean | 0.7982 |
| r2 std | 0.0491 |
| rmse mean | 0.8692 |
| rmse std | 0.1263 |
| q0 | -1.2855 |
| q1 | 1.4822 |
+-----------+---------+
A more precise tree model#
We conclude this part with the ExtraTreesRegressor
that seems to even even better performances.
from sklearn.ensemble import ExtraTreesRegressor
regressor = HistGradientBoostingRegressor()
model = make_pipeline(drop_all_nan_cols, simple_imputer, standard_scaler, regressor)
evaluation = evaluate_model(model, X, y)
result.append({'regressor': 'ExtraTrees'} | evaluation['metrics'])
evaluation['plot']
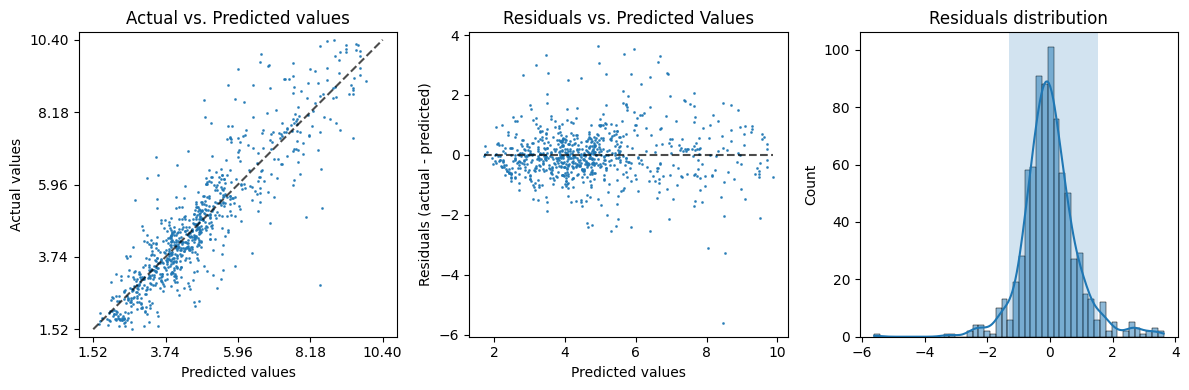
Quantitatively the results are even better:
print(evaluation['table'])
+-----------+---------+
| r2 mean | 0.7959 |
| r2 std | 0.0489 |
| rmse mean | 0.8745 |
| rmse std | 0.1293 |
| q0 | -1.3052 |
| q1 | 1.5198 |
+-----------+---------+
Choose, train and save the model#
We can put together the quantitative results to help us choose the best model:
from tabulate import tabulate
print(tabulate([r.values() for r in result], headers = result[0].keys(), tablefmt = 'outline', floatfmt = '.4f'))
+----------------------+-----------+----------+-------------+------------+---------+--------+
| regressor | r2 mean | r2 std | rmse mean | rmse std | q0 | q1 |
+======================+===========+==========+=============+============+=========+========+
| GradientBoosting | 0.8028 | 0.0333 | 0.8617 | 0.0931 | -1.2509 | 1.5986 |
| HistGradientBoosting | 0.7982 | 0.0491 | 0.8692 | 0.1263 | -1.2855 | 1.4822 |
| ExtraTrees | 0.7959 | 0.0489 | 0.8745 | 0.1293 | -1.3052 | 1.5198 |
+----------------------+-----------+----------+-------------+------------+---------+--------+
Suppose we choose the last (ExtraTrees) model, we can then train it with all the data at our disposal
from jp2rt import save_model
model.fit(X, y)
Pipeline(steps=[('columntransformer',
ColumnTransformer(remainder='passthrough',
transformers=[('drop_all_nan_cols', 'drop',
[216])])),
('simpleimputer', SimpleImputer()),
('standardscaler', StandardScaler()),
('histgradientboostingregressor',
HistGradientBoostingRegressor())])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('columntransformer',
ColumnTransformer(remainder='passthrough',
transformers=[('drop_all_nan_cols', 'drop',
[216])])),
('simpleimputer', SimpleImputer()),
('standardscaler', StandardScaler()),
('histgradientboostingregressor',
HistGradientBoostingRegressor())])ColumnTransformer(remainder='passthrough',
transformers=[('drop_all_nan_cols', 'drop', [216])])[216]
drop
[0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 141, 142, 143, 144, 145, 146, 147, 148, 149, 150, 151, 152, 153, 154, 155, 156, 157, 158, 159, 160, 161, 162, 163, 164, 165, 166, 167, 168, 169, 170, 171, 172, 173, 174, 175, 176, 177, 178, 179, 180, 181, 182, 183, 184, 185, 186, 187, 188, 189, 190, 191, 192, 193, 194, 195, 196, 197, 198, 199, 200, 201, 202, 203, 204, 205, 206, 207, 208, 209, 210, 211, 212, 213, 214, 215, 217, 218, 219, 220, 221, 222, 223, 224, 225, 226, 227, 228, 229, 230, 231, 232, 233, 234, 235, 236, 237, 238, 239, 240, 241, 242]
passthrough
SimpleImputer()
StandardScaler()
HistGradientBoostingRegressor()
Finally we can save the trained model using the save_model() function:
from jp2rt import save_model
save_model(model, 'extratrees')
315308
Use jp²rt#
Even if performing the above tasks allows for a more detailed understanding of model estimation and can be personalized with further processing and analysis steps to improve the quality of the model, sometimes can be a tedious and error prone process, especially for beginners.
To facilitate the process we can use the
simple_ensemble_model_estimate() function that takes as input
the name of a regressor, valid names are returned by the
list_ensemble_models() function:
from jp2rt import list_ensemble_models
list_ensemble_models()
['AdaBoost',
'Bagging',
'ExtraTrees',
'GradientBoosting',
'HistGradientBoosting',
'RandomForest']
For example, we can see what we get using AdaBoost:
from jp2rt import simple_ensemble_model_estimate
model = simple_ensemble_model_estimate('AdaBoost', X, y)
evaluation = evaluate_model(model, X, y)
evaluation['plot']
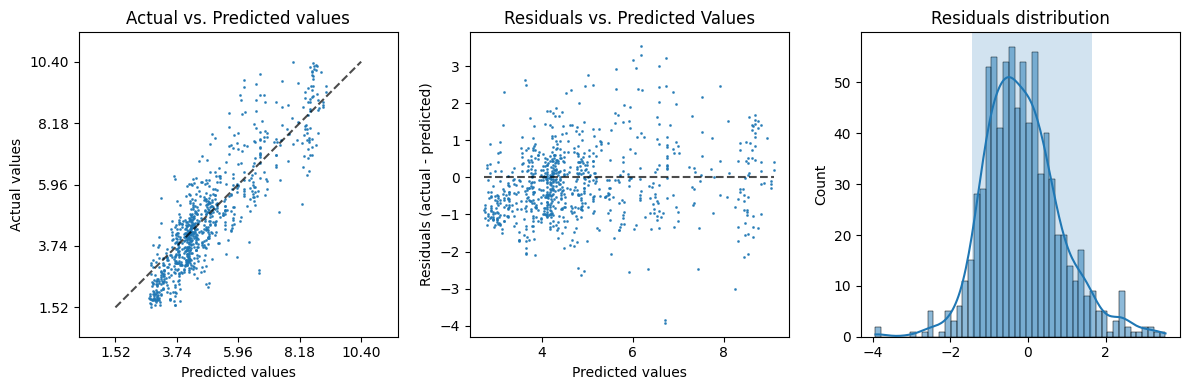
The command line#
To make things even easier, we can use the command line interface of jp²rt to list the available models:
$ jp2rt list-models
AdaBoost
Bagging
ExtraTrees
GradientBoosting
HistGradientBoosting
RandomForest
And to estimate (and evaluate) a model:
$ jp2rt estimate-model --evaluate Bagging docs/plasma+descriptors.tsv out
Read 799 molecules with 243 descriptor values each
Estimating model...
Model saved to /home/runner/work/jp2rt/jp2rt/docs/example/bagging.jp2rt (161647 bytes)...
Evaluating model...
+-----------+---------+
| r2 mean | 0.7758 |
| r2 std | 0.0286 |
| rmse mean | 0.9196 |
| rmse std | 0.0824 |
| q0 | -1.4328 |
| q1 | 1.5489 |
+-----------+---------+
Evaluation plot saved to /home/runner/work/jp2rt/jp2rt/docs/example/bagging.png
that will create the bagging.jp2rt containing the saved model, and
bagging.png the visual residual analysis.
